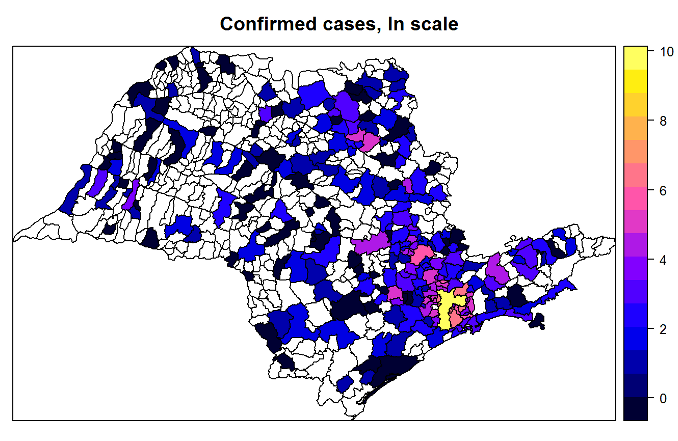
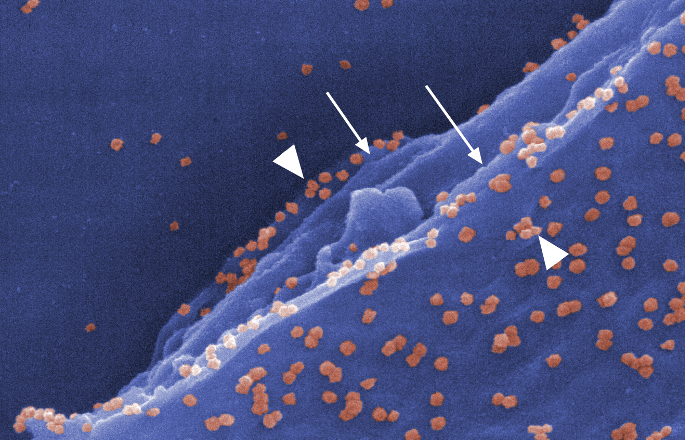
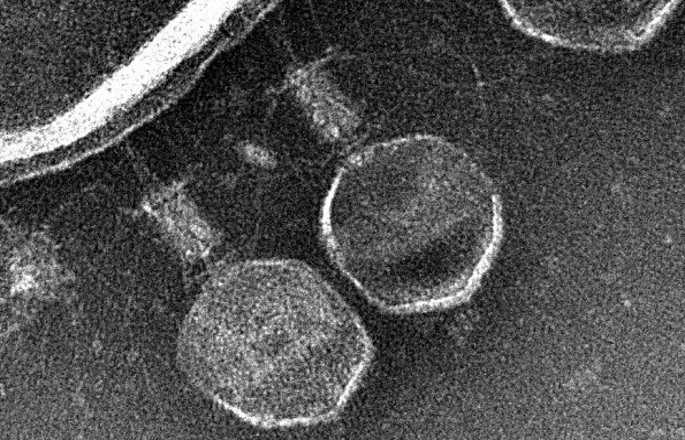
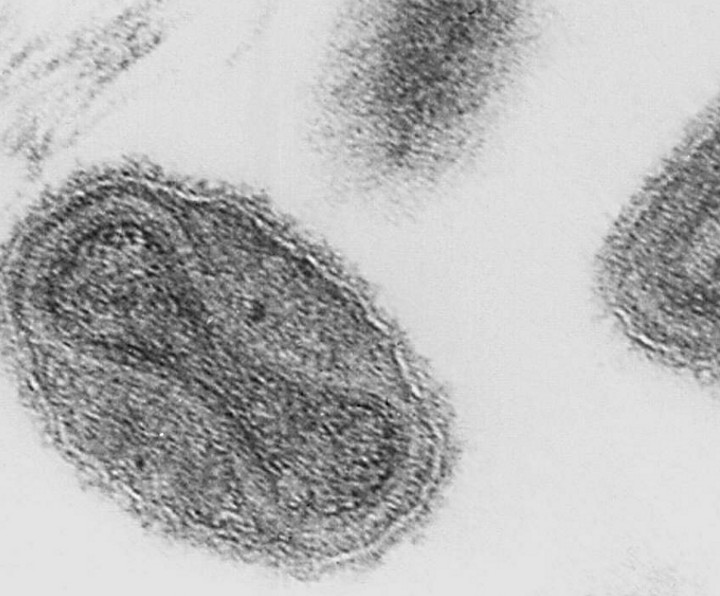
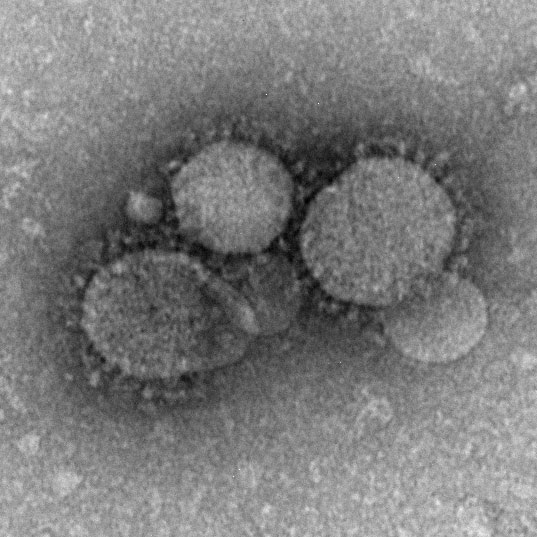
Superspreading in macroparasites
Maya Risin, a student from Emory University, worked with Dr. John Drake to study superspreading in macroparasites. Abstract: It is widely understood that host populations harboring macroparasites, which include parasitic helminths and arthropods, typically exhibit skewed infection burdens that give rise to “superspreading”. On the individual level, concurrent infections by multiple parasitic species within an individual