Mentor: Dr. Andrew Park group (http://parklab.ecology.uga.edu/)
Abstract: Viruses are notoriously capable of jumping between animal species. However, we’re only beginning to understand the roles of seasonality, contact probability, and host demography in this process. In Mongolia, there are as many horses as people, and they routinely get infected with avian influenza viruses as migrating birds stop over at water bodies in proximity to horses. Our group works with an international team to study bird to horse transmission of influenza in this region. This project will use the data we have collected to develop computational models describing transmission between host species. In particular, the goal is to better understand how season, horse proximity to birds, and horse age influence the probability of horses becoming infected.
Is the project computational, empirical, or both? Computational, including analysis of data and model building (Using R Studio & R Markdown)
Tag Archives: Quantitative
8) Detecting influenza A virus antigenicity with density-based algorithms
Mentor(s): Dr. Pej Rohani group
Abstract: Influenza viruses, such as H3N3 and H1N1, are a major cause of illness and death worldwide, accounting for over 3 million severe cases and approximately 500,000 deaths each year. These viruses also have a significant economic impact. The use of predictive modelling has been a useful in detecting new strains of the virus and developing the vaccines against the virus.
In this project, we will compare the predictive performance of density-based outlier detection algorithms, specifically the Empirical-Cumulative-distribution-based Outlier Detection (ECOD) algorithm, with isolation forest and one-class support vector machines (SVMs) in detecting unusual physicochemical properties in influenza A viruses. By identifying viruses with significantly different physicochemical properties, we can identify those that may be antigenically distinct, which has important implications for vaccine development and public health efforts.
We will use both simulated and benchmark data from Smith et al. for the experiments and fit the models using Python libraries such as Sciklearn, Pandas, Numpy, and PyoD. We will then use R packages, including ggplot2, ggdensity, dplyr, and tidyverse, for data visualization and analysis.
The ECOD algorithm has three desirable properties for this task: it can be fitted with limited hyperparameter tuning, it is computationally effective, and it improves interpretability. We will compare the predictive and inferential performance of all three algorithms. The results for isolation forest and one-class SVMs will be provided from previous experiments. We will end the project with a final report.
Is the project computational, empirical, or both? Computational.
7) Environmental variability and mosquito-borne disease
Mentors: Dr. Kyle Dahlin and Dr. John Vinson
Abstract: Climate change is expected to lead to shifts in the risk of mosquito-borne disease outbreaks throughout the world. However, it is unclear how increases in the variability of temperature and rainfall might impact these outbreaks. The goal of this project is to investigate how environmental and demographic noise affect key measures of the transmission of mosquito-borne diseases. The student will be responsible for conducting simulation experiments in R to explore the effect of noise on the probability of an outbreak and the outbreak size distribution. The student will gain experience in the ecology of vector-borne diseases, mathematical modeling, and programming in R.
Is the project computational, empirical, or both? Computational.
6) Development of infectious disease research and teaching software
Mentor(s): Dr. Andreas Handel group (http://handelgroup.uga.edu/)
Abstract: Using computer models to study infectious diseases can be challenging for students and researchers who are not trained as modelers. To make this process easier, we have developed several software packages, implemented in the popular R language, to help individuals learn about and analyze infectious disease models both at the individual and population level. The goal of this project is to further advance this software by implementing new features, making tutorials, testing existing features, adding new features, and more. This will increase the usefulness and power of the software and will give future students and researchers better tools to learn about and analyze different infectious diseases.
Is the project computational, empirical, or both? This project is computational. We will make use of the R language for all parts of this project.
5) Cataloging norovirus challenge study data
Mentor(s): Dr. Andreas Handel group (http://handelgroup.uga.edu/)
Abstract: A challenge study is a type of trial where patients are intentionally infected with a pathogen. Norovirus, everyone’s favorite cruise ship disease, has been studied in several challenge trials in the past, since its symptoms are relatively mild, and it is difficult to grow in the lab outside of live animals. While certain norovirus challenge studies are widely cited, there is no consensus among experts of exactly how many norovirus challenge studies have been conducted. We will review the published literature to find all previously published challenge studies, and where possible, extract the data from these studies in order to gain a more comprehensive understanding of how many norovirus challenge trials have been conducted and what information they have provided.
Is the project computational, empirical, or both? This project is computational and will use the R programming language where necessary. We do not expect this project to be programming intensive.
4) Modelling outcomes of influenza infection and vaccination
Mentor(s): Dr. Andreas Handel group (http://handelgroup.uga.edu/)
Abstract: Everyone responds differently to the flu shot – the vaccine does not provide 100% protection like some other diseases, and the vaccine needs to be updated every year because flu is able to rapidly evolve and quickly evade immune defenses. The immunological mechanisms for these differences are still not well understood, despite years of previous scientific work. As part of a multi-center collaboration, we will combine data from several studies of seasonal influenza vaccines, and hunt for clues. By leveraging the power of data visualization, machine learning, and statistical modeling techniques, we can identify which aspects of a person, vaccine, or flu strain might affect an individual’s response to the flu vaccine.
Is the project computational, empirical, or both? This project will be entirely computational, and will consist of data exploration and statistical analyses. We will use the R programming language, and this project will involve a substantial amount of programming.
3) Understanding spatiotemporal dynamics of chronic wasting disease in white-tailed deer
Mentors: Dr. Elizabeth M. Warburton and Marcelo Jorge
Abstract: Since it was first detected in free-ranging elk in the USA in 1981, chronic wasting disease (CWD) has spread to multiple cervid species, including moose, mule deer, and white-tailed deer, across 30 states. In some locations where this deadly disease has spread, more than 50% of the local deer population is infected. CDW is a prion disease, or transmissible spongiform encephalopathy (TSEs), caused by infectious proteins similar to those that cause bovine spongiform encephalopathy (“mad cow disease”) and Creutzfeldt-Jakob disease in humans. Like all TSEs, CWD is a progressive, fatal disease that affects the brain, spinal cord, and many other tissues. Unlike some other TSEs, CWD can be either directly transmitted by close contact between hosts or environmentally transmitted. The combination of these prions possibly remaining infectious in soil for years and the approximately year-long asymptomatic period in hosts makes understanding the spread of this disease through the white-tailed deer population especially challenging. We use camera-trap, telemetry, and capture-mark-recapture data from our study sites in NW Arkansas to understand the spatiotemporal dynamics of CWD infection in white-tailed deer. By using computational methods such as Bayesian hierarchical models, individual-based models, and machine learning we seek to characterize and predict disease spread. We then communicate our findings to conservation professionals so they can better manage this deadly disease in free-ranging cervids.
Is the project computational, empirical, or both? This project is mainly computational but also uses field-collected data. Students will be involved in analyzing camera-trap, telemetry, and capture-mark-recapture data as well as creating simulations of disease spread.
2) How do predators affect disease dynamics in their prey? Experimental tests of the healthy herds hypothesis with fish predators, zooplankton hosts, and a fungal parasite
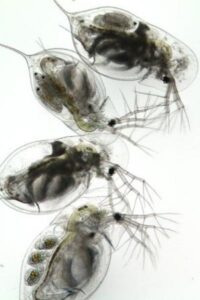
Mentors: Dr. Robbie Richards and Dr. Alex Strauss
Abstract: Food web members can dramatically impact host-parasite dynamics through a wide variety of mechanisms. The “healthy herds hypothesis” posits that predators can substantially decrease parasitism in their prey by directly consuming infected individuals. However, experimental tests of this idea remain rare. Moreover, the few experiments that have been attempted have yielded inconsistent results, with predators sometimes decreasing disease, increasing disease, or having no effect on disease in their prey. Reconciling these divergent outcomes is increasingly important as prey species serve as reservoirs for many diseases of concern for spillover into human populations. In this project you will have the opportunity to conduct a large-scale manipulative experiment to test the healthy herds hypothesis. We will use a replicated mesocosm (i.e., artificial pond) system of mosquitofish predators, fungal parasites and their shared host/prey, the water flea, Daphnia dentifera. Finally, we will measure several key traits (e.g., preference of fish for infected vs. healthy hosts) and ask whether they explain why predators increase, decrease, or have no effect on disease in the experiment.
Is the project computational, empirical, or both? Mostly empirical, but with opportunities to learn computational skills such as model fitting and parameterization.
How Various Feeding Rates Affect Pupation Rates in Anopheles stephensi Larvae
Jacob Glover, a student at Franklin College, worked in the lab of Dr. Ash Pathak
Abstract Anopheles Stephensi are a dangerous vector for countless diseases without cures currently. If mosquitos could be contained or controlled, then this could eliminate these diseases they carry without having to find a vaccine or other types of cures for them. One of the best stages to control mosquitos is within their larval stage of life. In this stage, they are so fragile that minor changes in food or environment can kill them or stunt their growth. If mosquito larvae could be purposefully stunted or killed off, then there would be one less major vector for disease. Our experiment tests this hypothesis to see if manipulating the rate at which mosquito larvae are fed if that impacts their pupation rates.Within this experiment, we test to see if various feeding rates affect their pupation rates, ultimately testing if their growth stays the same under different feeding conditions. This was done by giving them different amounts of TetraFin pellets on Tuesdays and Wednesdays to see if there was a difference in growth rates. Overall, the results show that no significant changes occur in the pupation rates of mosquito larvae when feeding rates are altered.
Glover_posterModeling Fitness of Immune Evading Pertussis Mutants
Gowri Vadmal, a student at Stanford University, worked in the lab of Dr. Pej Rohani
Abstract Pertussis was considered one of the great diseases of childhood with most people experiencing a bout of the infection by the age of 15. The initial roll out of vaccines in the 1950s led to a marked decline in pertussis incidence, with optimism over its potential elimination. Over the past 20-30 years, however, a clear increasing trend in pertussis cases has emerged. A number of putative Pertussis (whooping cough) is caused mainly by the bacterium Bordetella pertussis. Many countries use acellular pertussis vaccines containing the antigen pertactin (PRN), which plays an important role in pathogenesis. In recent years, we’ve observed an increasing number of B. pertussis isolates that are PRN-deficient and able to infect people even in highly vaccinated countries. We used SIRV models to look at the fitness cost of the bacterium losing PRN and the advantage of being able to infect already vaccinated people. Strains can invade the population only if their leakiness (ability to infect vaccinated people) and transmission are above the threshold for invasion, which depends on vaccination coverage and the cost of immune evasion for the strain. At low vaccination coverage, strains with high leakiness dominate the system when the fitness cost to evade immunity is low, but as cost increases for the strains to infect people, there is bistability between the wildtype and mutant strains. However, at higher vaccination coverage, the wild type completely fades out and the strains with the highest leakiness dominate the system. Thus, the conditions for B. pertussis mutant invasion can change depending on a population’s vaccine coverage, the cost of losing the pertactin gene, and the advantage of being able to evade vaccine immunity.
Vadmal_posterDesigning and Evaluating the Need for Patient Based Clinical Prediction Rules for Influenza Triage Telemedicine
Annika Cleven, a student at St. Olaf College, worked in the lab of Dr. Andreas Handel
Abstract Using data that was collected from a university health center where patients and clinicians were asked to report the presence of a list of respiratory-related symptoms, we analyzed the need for patient based Clinical Prediction Rules (CPRs). We investigated inter-rater agreement between clinicians and patients on symptom reporting and found they often disagreed. A Bland-Altman analysis indicated that the disagreement on symptom reporting was enough to invalidate using patient reported symptom data on a CPR that was designed using clinician reported data. Further analysis determined that using patient versus clinician reported symptom data could lead to different risk category designations and advised care. In response, we built risk prediction models that were fit to patient reported data and they provided minimal improvement from the clinician based CPRs. Our research indicates that clinician based CPRs cannot be effectively used with patient reported symptoms data, like in telemedicine. Our results also show that we can create a patient-based CPR, but because of the minimal improvement, a combination of at-home testing and a patient-based CPR would be the best approach to influenza triage telemedicine.
Cleven_posterDeforestation alters spillover risk of multi-host pathogens
Annalise Cramer, a student at Westfield State University, worked in the lab of Dr. Richard Hall
Abstract Deforestation alters landscape configuration resulting in novel contacts between host species, which can promote pathogen spillover from wildlife to domesticated animals and humans. Given heightened awareness of zoonotic spillover, studies are urgently needed to understand how the rate of deforestation interacts with host abundance and distribution to shape pathogen transmission across habitats and human exposure risk. In this study, we derive a mathematical model coupling land use change with pathogen transmission between hosts in forested and deforested habitats. We explore how deforestation rate and host relative abundance across habitats influence the dynamics, peak and cumulative number of infected hosts in deforested habitats as a proxy for human spillover risk. We find that the number of infected hosts in deforested habitats peaks sooner under faster deforestation rates. When the deforested hosts are less abundant, most transmission occurs in mixtures of forested and deforested habitats where large habitat boundaries maximize contacts with the more abundant forest hosts. This results in a hump-shaped relationship between deforestation rate and short- and long-term spillover risk. These results suggest that surveillance and interventions at habitat boundaries are crucial to reduce the risk of zoonotic spillover.
Cramer_posterOrNet: Spatiotemporal Analysis of Organelle Morphology
Walter Avila, a student from Emory University, worked in the lab of Dr. Shannon Quinn.
Abstract Modeling changes in organelle morphology in response to infections is pivotal in studying pathogenic behaviors. Mitochondria are the most meaningful organelles to study because their structure changes dramatically in the presence of potentially fatal infections. Accurately modeling mitochondria could provide crucial information about severe infections. The project addresses this modeling concern by continuing development on OrNet (Organellar Networks). This Python framework constructs dynamic social network graphs of fluorescently tagged cells in microscopy videos to analyze the spatiotemporal relationships of mitochondria. These graphs are leveraged into eigenvalue arrays to describe organelle morphology over time quantitatively. We sought to improve the Temporal Anomaly Detection (TAD) technique in OrNet, which utilizes the arrays and detects when abnormal events occur in a video, indicative of organellar changes. The number of arrays rows and columns corresponds to the number of frames and eigenvalues, respectively. Currently, TAD takes the average eigenvalue at each frame and computes the distance between the average and the mean of a few preceding averages. If that distance exceeds a threshold, the frame is an outlier. This parameter-sensitive technique gives disproportionate weight to trailing eigenvalues. We assess how differently this model behaves when introducing a weighted average eigenvalue per frame. Using live videos of HeLa cells in different mitochondrial conditions, on average, the weighted average TAD declares a higher proportion of anomalous frames in cell videos from each mitochondrial condition than the simple average TAD code. This information about our model will guide how we implement OrNet in the future. Comparing how videos are analyzed differently by both TAD versions and other comparisons are necessary. We ultimately aim to use OrNet in large-scale genomic screenings of Mycobacterium tuberculosis mutants to understand better how these pathogens invade cells and induce cell death at the genetic level.
AvilaFrugivory Richness Predicts Ebola Spillover in Africa
Mireya Dorado, a student at Northeastern University, worked in Dr. Patrick Stephen’s lab studying pathogen spillover
Ebola is a deadly filovirus that infects a variety of mammals including humans. Since the first documented outbreak i n 1976, there have been numerous field studies searching for the source of the spillover of Ebola. Only a few studies have directly investigated the effect of mammalian host biodiversity. These studies have been limited to the diversity of known Ebola hosts and bats. However due to Ebola’s broad host range, there has not been a systematic approach to which hosts may be important for spillover. Therefore, our goal was to determine whether and what aspects of mammalian diversity play a significant role in predicting Ebola spillover events. We calculated species richness of mammals in 50 kilometer by 50 kilometer grid cells across Africa. Statistical analyses were based on a presence absence approach, which compared species richness at sites of spillover t o pseudo-absence background locations. We used bagged logistic regression, a machine learning method, to create statistical models testing how well species richness of different mammal subgroups predicted spillover. Overall, we found that Cercopithecidae and Pteropodidae were the strongest taxonomic predictors of spillover (mean AUC=0.943 and 0.936 respectively), but diversity of frugivorous species was the best overall predictor (mean AUC=0.956). This strongly implicates a role of fruit in Ebola transmission and the significance of fruiting and masting seasons as ideal times for spread of infection.
DoradoInvestigating the Clinical Relevance of Patient-Reported Symptoms for Influenza Triage
Jacqueline Dworaczyk, a student at Arizona State University, worked in the lab of Dr. Andreas Handel.
Telemedicine has become increasingly popular during the age of Covid-19. During a public health crisis, telemedicine could be used as a tool to triage patients and prevent burden on the health care system. In an exploratory data analysis, we investigated whether a symptom questionnaire could be used to predict influenza diagnosis. A symptom questionnaire containing 19 upper respiratory symptoms was administered to patients and clinicians (n = 2475 patients) at University of Georgia’s Health Center during the 2016-2017 flu season. Five clinical decision rules were applied to the symptoms reported by the patients and clinicians. The clinical decision rules’ performance when predicting influenza diagnosis was assessed using AUC, F1, MCC, sensitivity and specificity. A 7-11% drop in AUC was observed across all clinical decision rules when using the patient-reported symptoms as opposed to the clinician-reported symptoms.
In a sensitivity analysis, we evaluated the clinical decision rules’ ability to predict true, lab-confirmed influenza in a subset of our population who received PCR testing. While clinician-reported symptoms still performed better than patient-reported symptoms, the difference in AUCs when predicting PCR was significantly smaller. These differences in performance may be partially explained by a lack of agreement between patients and clinicians on the presence of signs and symptoms. Agreement between the patients and clinician’s questionnaire responses (n = 2475) was quantified using Cohen’s Kappa statistic and found that at best, patient’s and clinician’s had moderate agreement on three of the nineteen symptoms assessed. Overall, it was found that using a patient symptom questionnaire to predict physician diagnosis led to a reduction in accuracy. Further studies need to be done to assess the clinical relevance of this reduction.
DworaczykInfectious disease professionals need better training in modeling
Salil Goyal, a student at the University of California Berkeley, worked with Dr. Andreas Handel.
Models have become increasingly important in the field of infectious disease epidemiology, and broadly in the field of public health, in recent years because the ability of scientists and officials to make educated decisions based on data is important. However, many authors have stated recently (and especially during COVID) that programs that train people working with infectious diseases generally do not impart adequate training in understanding models. While many resources aimed at teaching infectious disease modeling do exist, they are aimed at different audiences and there currently exists no synthesis of these resources. As a potential solution to the problem, we offer here a comprehensive review of resources relevant to the pedagogy of infectious disease epidemiology, with a focus on modeling.
GoyalIs relative viral load an important metric for treatment and prognosis of influenza A?
Zane Billings, a student at Western Carolina University, worked with Dr. Andreas Handel and graduate student Brian McKay in the UGA College of Public Health.
Abstract
Influenza-like illnesses (ILIs) present with several of the same symptoms, including cough, fatigue, and weakness. However, ILIs can be caused by a range of different pathogens with vastly different treatments. Quantitative PCR is an incredibly specific and sensitive method to detect several ILIs, but until recently, qPCR methods were prohibitively expensive and required special training and equipment. Recent advances in qPCR technology have allowed for machines such as the Roche cobas Liat system to become available to point-of-care physicians. Using data collected from the University of Georgia Student Health Center, qPCR data was examined relative to patient and physician reported symptoms, as well as impacts and recovery from disease to determine if quantitative estimates of relative viral load are important for physicians to make informed decisions. While relative viral load estimates were found to be correlated to days since the onset of illness and patient temperature at diagnosis, no correlations were found between recovery or severity of illness and relative viral load. However, the study sample was very limited and more research should be performed on broader study populations.
Understanding the dynamics of viral shedding within Norovirus infected subjects
Simran Budhwar from the University of Virginia, worked with Rachel Mercaldo, Brian McKay, and Dr. Andreas Handel to study shedding of Norovirus.
Abstract: Norovirus (NoV) is a common cause of acute gastroenteritis. Symptoms include vomiting and diarrhea, which can lead to complications such as dehydration and also serve to spread viral particles through bodily fluids. While some infections are asymptomatic, infected individuals shed the virus regardless of disease severity, primarily through stool. To understand the dynamics of viral shedding, previous studies measured viral load in healthy human subjects challenged with various doses of the virus. In the present analysis, data from these studies was combined to better describe NoV shedding over time. We calculated key variables such as peak viral titer, time to peak viral titer, and duration of shedding, in addition to estimating total shedding through the area under the curve (AUC) value of each participant’s total shedding time-series curve. On average, patients shed the virus for 22 days, with the peak viral titer appearing on day 5 following challenge. Peak viral titers were 10.551 (log10) genomic equivalence copies per gram of stool, while AUC averaged at 11.58 (log10) genomic equivalence copies per gram stool. Though these are key variables that are necessary to understand viral shedding, future work should focus on exploring the drivers of variation in viral load and shedding, such as symptoms or other patient-specific factors.
BudwharGenomics of bacterial symbionts to determine nutritional roles in plant-sap feeding insects
Michael Lansford, a student at the University of Rochester, worked with Dustin Dial and Dr. Gaelen Burke
Abstract: Adelgids are sap-sucking insects that contain bacterial endosymbionts to help them synthesize essential amino acids. The adelgid life cycle alternates between sexual generations that parasitize spruce as a primary host and asexual generations that parasitize a secondary host plant species. The adelgid family consists of five lineages each with a different secondary host plant: Douglas fir, fir, hemlock, larch, and pine. Each adelgid lineage has a different pair of symbionts, a primary symbiont that was acquired by the adelgid first and a secondary symbiont that was acquired second. Vallotia is a symbiont shared between the Douglas fir lineage, where it is the secondary symbiont, and the larch lineage, where it is the primary symbiont. To determine the nutritional roles of Vallotia in different species, genomic data were searched for genes involved in amino acid synthesis. FastQC was used to evaluate the quality of raw adelgid read data. The Georgia Advanced Computing Research Center (GACRC) cluster was used to assemble and annotate genomes from the raw reads. After running scripts to assemble raw reads into scaffolds, BLAST was used to identify which scaffolds were from symbionts. Symbiont genes were annotated using PROKKA and Geneious Prime and biochemical pathways were reconstructed with help from BioCyc. The results showed that Vallotia is primarily responsible for synthesis of all essential amino acids except cysteine in the Douglas fir lineage species A. cooleyi. Gillettellia, the primary symbiont in the Douglas fir lineage, works together with Vallotia in lysine and aromatic amino acid synthesis. In both larch lineage species, Vallotia is only responsible for the final steps in tryptophan synthesis and depends on the secondary symbiont Profftia in A. lariciatus and probably A. abeitis for most steps in aromatic synthesis. These results suggest that Vallotia was acquired by the Douglas fir lineage to account for the loss of most synthesis genes in Gillettellia and Profftia was acquired by the larch lineage to account for the loss of aromatic synthesis genes in Vallotia.
Lansford_newTo skip or not to skip: exploring the connections between orviposition behavior and density –dependence in Aedes albopictus mosquitoes
Taryn Waite, a student at Colby College, collaborated with REU student Courtney Schreiner, Nicole Solano, Dr. Craig Osenberg, and Dr. Courtney Murdock.
Abstract: Conspecific density in larval habitats is an important factor affecting adult fitness in Aedes albopictus mosquitoes, as it drives competition for food and space. We conducted a larval density experiment wherein mason jars containing leaf infusion and varying numbers of larvae were placed in a field enclosure, developmental stage was recorded daily, and emerged adults were collected. Nonlinear regressions were performed on the data for survival to adulthood, sex ratio of adults, and wing length of females, and fecundity was inferred from wing length. Using these regressions, an equation was created to predict short-term population dynamics in habitats with varying conspecific densities. What determines the densities that will actually occur in various larval habitats is where females choose to lay their eggs. Female mosquitoes have the ability to skip-oviposit, which entails spreading their eggs out among multiple habitats instead of dumping them all in one habitat. The population dynamics equation was used to evaluate the theoretical consequences of skip- versus non-skip- oviposition, using scenarios with varying numbers of egg-laying females and a fixed number of available larval habitats. We found that at low densities of ovipositing females, skip-oviposition produces more short-term population growth than non-skip-oviposition. At higher densities, non-skipping becomes more productive than skipping, though there is less divergence between the outcomes. This simulation demonstrates a way in which patterns of density-dependence could act as a link between oviposition behavior and population dynamics. Due to the effects that we found of density dependence in larval habitats, individual females’ oviposition behavior could have consequences for short-term population dynamics.
Waite

